Current Projects
Project 1: Developing new antibiotics by targeting specific molecular interactions in bacteria
Antibiotic resistance is an increasing problem globally and may bring about the end of modern medicine as we know it
Predictions suggest the number of death from drug-resistant infections will surpass those from cancer by 2050
New antibiotics are urgently required but their discovery has slowed dramatically in the last two decades
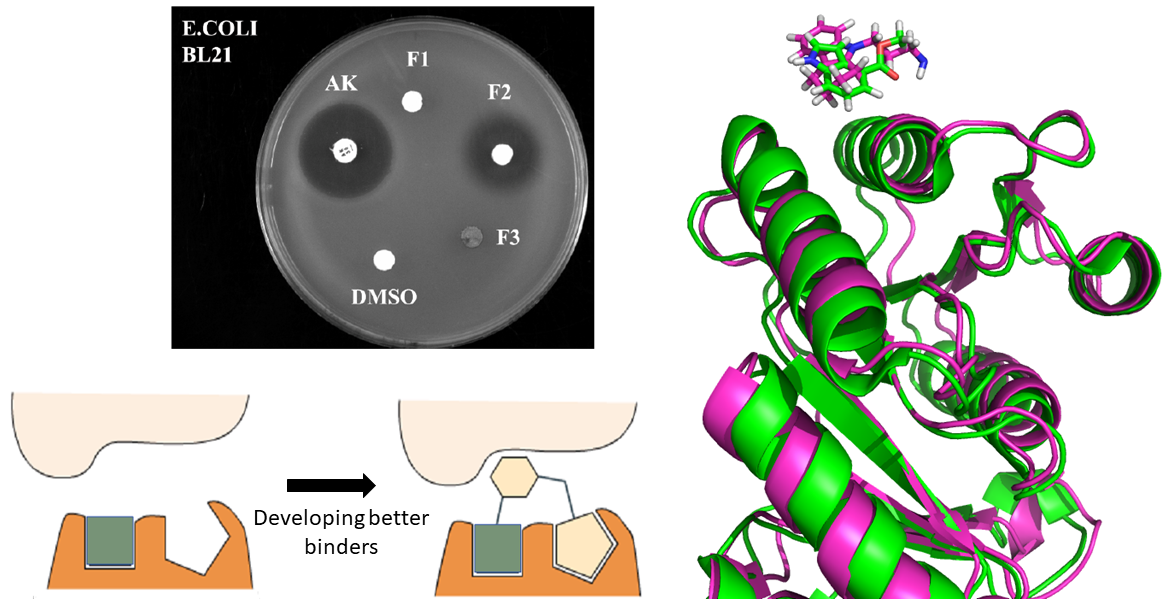
We are developing new antibiotics by targeting a never exploited interaction unique to bacteria using a number of approaches including fragment-based drug design (FBDD), engineered nucleic acid mimetics.
This work is a collaboration with Dr Lorna Wilkinson-White and Dr Toby Passioura (Sydney Analytical) and A/Prof Belinda Abbott (La Trobe) and others.
Techniques used in this project: antimicrobial assays, fragment and structure-based drug design, X-ray crystallography, protein biophysics, in vitro evolution
Project 2: Targeting protein:RNA interactions to fight COVID
Despite the availability of vaccines, there is still an unmet need for effective drugs to treat COVID
Critical protein:RNA interactions in SARS-CoV2 will be targeted in this project and techniques used will be similar to Project 1
This is a new project, so another focus is to characterise critical protein:RNA interactions in SARS-CoV2
This work is a collaboration with Dr Sandro Ataide, Dr Biswaranjan Mohanty (Sydney Analytical) and Dr Roland Gamsjaeger (Western Sydney University).
Techniques used in this project: in vitro evolution of peptides, structure-based drug design, X-ray crystallography, protein biophysics
Project 3: Engineering of hydrophobins for medical implants
Medical implant infections account for ~45% of hospital-acquired infections
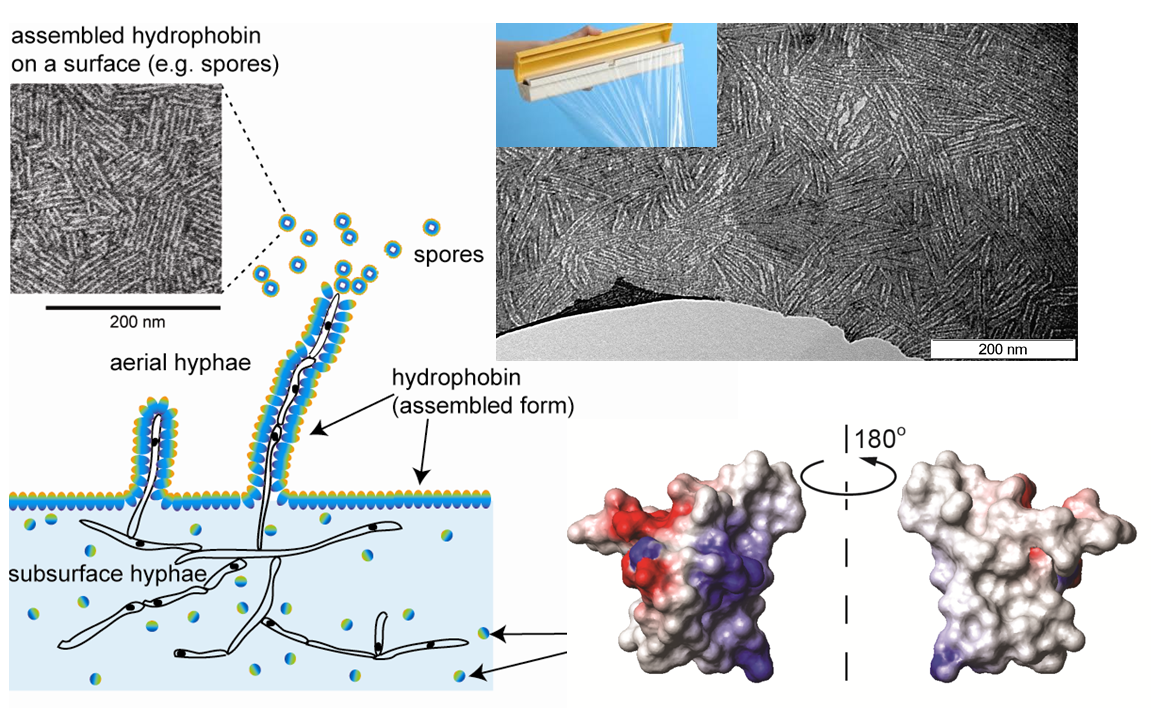
Hydrophobins are fungal proteins that spontaneously self-assemble at interfaces to form robust and amphipathic coating
Hydrophobin coatings have been shown to improve biocompatibility of hydrophobic surfaces
Can we expand the functionality of hydrophobin coatings through protein engineering to include antibacterial properties?
Together with researchers at School of Medical Science (Prof Margie Sunde), School of Physics (Prof David McKenzie and A/Prof Natalka Suchoworska) and the Chris O’brien Lifehouse, we are trying to engineer hydrophobins that can promote the adhesion and growth of osteoblasts while preventing infections in PEEK bone implants.
Techniques used in this project: protein biochemistry, high-resolution imaging, protein biophysics and engineering, cell culture assays
Project 4: Using omics to detect infectious diseases and to evaluate new cancer therapies
This project has two directions:
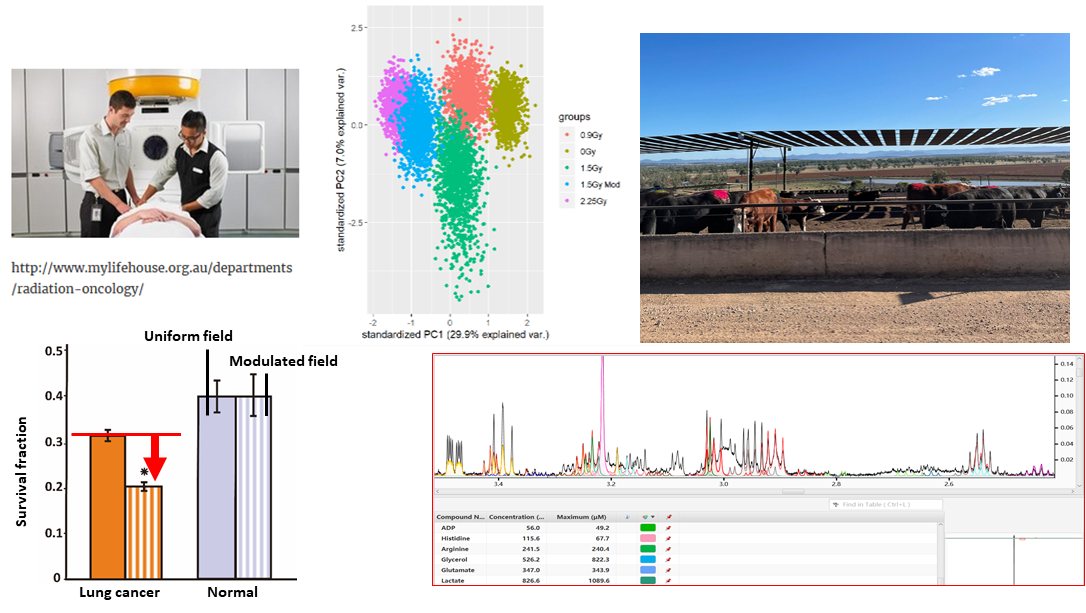
The use of mass spectrometry and NMR experiments to identify and quantify metabolites that can act as early predictors and biomarkers of respiratory diseases in cattle. This would not only reduce cattle mortality and morbidity but also reduce the overuse of antibiotics in agriculture. This project is a collaboration with Prof Luciano Gonzalez.
Using a combination of metabolomics and transcriptomics to evaluate new cancer therapies from new radiotherapy schemes to physical plasma treatment. This project is a collaboration with School of Physics (Prof David McKenzie and A/Prof Natalka Suchoworska) and the Chris O’brien Lifehouse.
Techniques used in this project: NMR spectroscopy, mass spectrometry, metabolomics, transcriptomics, cell culture, biochemical assay development
Project 5: Developing specific inhibitors that target periodontal diseases
Porphyromonas gingivalis is a “keystone” pathogen in chronic periodontitis which affects a quarter of the world’s populationexploiting the specific pathogenic traits of bad bacteria such as Porphyromonas gingivalis. This is narrow-spectrum antibiotic project is co-supervised by Dr Jinlong Goa, Sydney Dental School
Symptoms include inflammation and bleeding which eventually lead to destruction of tooth and supporting structures
We have identified several proteins including Haem uptake system protein A (HusA) and a novel protein which we have termed FetB which are associated with P. gingivalis becoming virulent and enable growth under iron-restricted conditions, e.g. in plaques
We have determined the structure of HusA and FetB and performed docking studies with haem and analogues. This has allowed us to design and test the idea of “Trojan horse” inhibitors for HusA. We are working on improving the specificity and potency of the lead compounds.
Docking studies showing how haem and a prototype “Trojan” inhibitor bind to HusA
Techniques used in this project: protein chemistry, NMR spectroscopy, mass spectrometry, inhibitor design, biological assays